Computational Analysis of N-ferrocenylmethyl-N-phenylbenzohydrazide: Molecular Docking and Dynamic Stability with BSA
Abstract
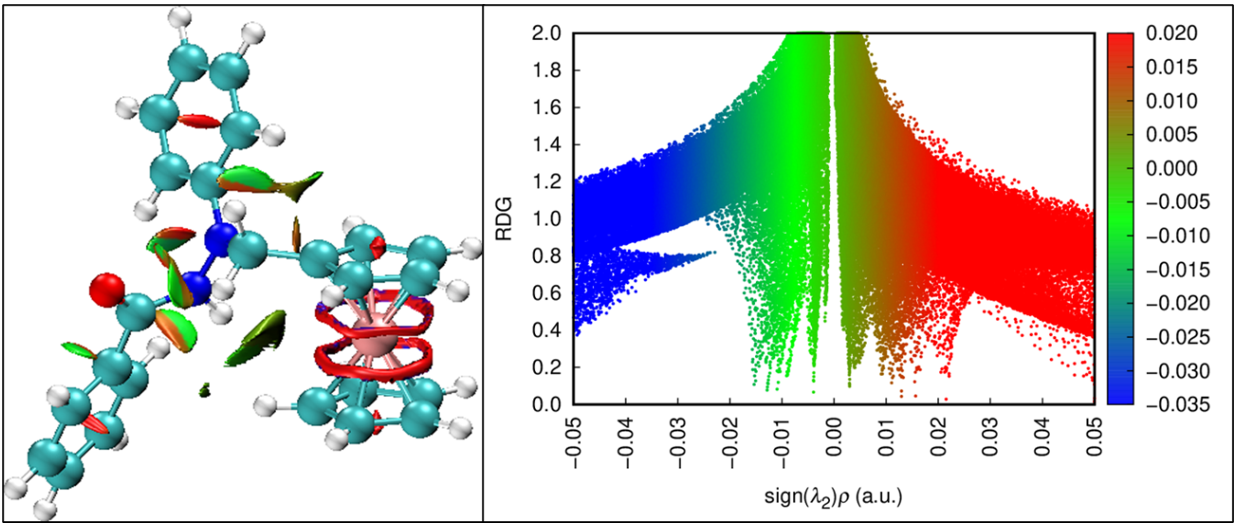
This study provides a comprehensive computational analysis of N-ferrocenylmethyl-N-phenylbenzohydrazide (FHD) as a potential bioactive compound by employing density functional theory (DFT), molecular docking, and molecular dynamics (MD) simulations. DFT calculations, including frontier molecular orbital (FMO) and molecular electrostatic potential (MEP) analyses, reveal the electronic structure and reactive regions of FHD. Reduced density gradient (RDG) analysis highlights weak intermolecular interactions, confirming molecular stability. Molecular docking studies show strong binding of FHD to bovine serum albumin (BSA, PDB ID: 6QS9), with a binding energy comparable to the anti-inflammatory drug diclofenac (DIF). MD simulations demonstrate that the 6QS9-FHD complex exhibits superior stability over 100 ns, lower root mean square deviation (RMSD), and consistent compactness compared to 6QS9-DIF. Radius of gyration (Rg) and solvent-accessible surface area (SASA) analyses further confirm the stability of the FHD complex. These findings suggest FHD’s potential as a candidate for anti-inflammatory applications.
Downloads
References
M. Zloh, S.B. Kirton, The Benefits of In Silico Modeling to Identify Possible Small-Molecule Drugs and Their Off-Target Interactions, Future Med Chem 10 (2018) 423–432. https://doi.org/10.4155/fmc-2017-0151.
S. Tian, J. Wang, Y. Li, D. Li, L. Xu, T. Hou, The application of in silico drug-likeness predictions in pharmaceutical research, Adv Drug Deliv Rev 86 (2015) 2–10. https://doi.org/10.1016/j.addr.2015.01.009.
A. Villalobos, M. Welch, J. Minshull, In Silico Design of Functional DNA Constructs, in: 2012: pp. 197–213. https://doi.org/10.1007/978-1-61779-564-0_15.
V.L. Maruthanila, R. Elancheran, N.K. Roy, A. Bhattacharya, A.B. Kunnumakkara, S. Kabilan, J. Kotoky, In silico Molecular Modelling of Selected Natural Ligands and their Binding Features with Estrogen Receptor Alpha, Curr Comput Aided Drug Des 15 (2018) 89–96. https://doi.org/10.2174/1573409914666181008165356.
F.O. Vannberg, S.J. Chapman, A.V.S. Hill, Human genetic susceptibility to intracellular pathogens, Immunol Rev 240 (2011) 105–116. https://doi.org/10.1111/j.1600-065X.2010.00996.x.
E. Janik, M. Ceremuga, M. Niemcewicz, M. Bijak, Dangerous Pathogens as a Potential Problem for Public Health, Medicina (B Aires) 56 (2020) 591. https://doi.org/10.3390/medicina56110591.
C.A. Dinarello, Proinflammatory Cytokines, Chest 118 (2000) 503–508. https://doi.org/10.1378/chest.118.2.503.
R.H. Straub, C. Schradin, Chronic inflammatory systemic diseases – an evolutionary trade-off between acutely beneficial but chronically harmful programs, Evol Med Public Health (2016) eow001. https://doi.org/10.1093/emph/eow001.
X.-F. Huang, L.-Z. Wang, L. Tang, Y.-X. Lu, F. Wang, G.-Q. Song, B.-F. Ruan, Synthesis, characterization and antitumor activity of novel ferrocene derivatives containing pyrazolyl-moiety, J Organomet Chem 749 (2014) 157–162. https://doi.org/10.1016/j.jorganchem.2013.08.043.
J.-N. Wei, Z.-D. Jia, Y.-Q. Zhou, P.-H. Chen, B. Li, N. Zhang, X.-Q. Hao, Y. Xu, B. Zhang, Synthesis, characterization and antitumor activity of novel ferrocene-coumarin conjugates, J Organomet Chem 902 (2019) 120968. https://doi.org/10.1016/j.jorganchem.2019.120968.
B. Lal, A. Badshah, A.A. Altaf, M.N. Tahir, S. Ullah, F. Huq, Synthesis, characterization and antitumor activity of new ferrocene incorporated N,N′-disubstituted thioureas, Dalton Transactions 41 (2012) 14643–14650. https://doi.org/10.1039/c2dt31570j.
B. Lal, A. Badshah, A.A. Altaf, N. Khan, S. Ullah, Miscellaneous applications of ferrocene‐based peptides/amides, Appl Organomet Chem 25 (2011) 843–855. https://doi.org/10.1002/aoc.1843.
F. Asghar, A. Badshah, B. Lal, S. Zubair, S. Fatima, I.S. Butler, Facile synthesis of fluoro, methoxy, and methyl substituted ferrocene-based urea complexes as potential therapeutic agents, Bioorg Chem 72 (2017) 215–227. https://doi.org/10.1016/j.bioorg.2017.04.016.
J.C. Swarts, E.W. Neuse, G.J. Lamprecht, Synthesis and characterization of water-soluble polyaspartamide-ferrocene conjugates for biomedical applications, Journal of Inorganic and Organometallic Polymers 4 (1994) 143–153. https://doi.org/10.1007/BF01036539.
D.R. van Staveren, N. Metzler-Nolte, Bioorganometallic Chemistry of Ferrocene, Chem Rev 104 (2004) 5931–5986. https://doi.org/10.1021/cr0101510.
T. Itoh, S. Shirakami, N. Ishida, Y. Yamashita, T. Yoshida, H.S. Kim, Y. Wataya, Synthesis of novel ferrocenyl sugars and their antimalarial activities., Bioorg Med Chem Lett 10 (2000) 1657–9. https://doi.org/10.1016/s0960-894x(00)00313-9.
S. Benabdesselam, H. Izza, T. Lanez, E.K. Guechi, Synthesis, antioxidant and antibacterial activities of 3-nitrophenyl ferrocene, IOP Conf Ser Mater Sci Eng 323 (2018) 012007. https://doi.org/10.1088/1757-899X/323/1/012007.
C. Biot, N. François, L. Maciejewski, J. Brocard, D. Poulain, Synthesis and antifungal activity of a ferrocene-fluconazole analogue., Bioorg Med Chem Lett 10 (2000) 839–41. https://doi.org/10.1016/s0960-894x(00)00120-7.
B. Long, C. He, Y. Yang, J. Xiang, Synthesis, characterization and antibacterial activities of some new ferrocene-containing penems, Eur J Med Chem 45 (2010) 1181–1188. https://doi.org/10.1016/j.ejmech.2009.12.045.
S. Ali, A.A. Altaf, A. Badshah, Imtiaz-Ud-Din, B. Lal, S. Kamal, S. Ullah, DNA interaction, antibacterial and antifungal studies of 3-nitrophenylferrocene, Journal of the Chemical Society of Pakistan 35 (2013) 922–928.
E.M. Njogu, B. Omondi, V.O. Nyamori, Synthesis, physical and antimicrobial studies of ferrocenyl-N-(pyridinylmethylene)anilines and ferrocenyl-N-(pyridinylmethyl)anilines, South African Journal of Chemistry 69 (2016) 51–66. https://doi.org/10.17159/0379-4350/2016/v69a7.
A. Adaika, A. Adaika, T. Lanez, E. Lanez, in Vitro and in Silico Evaluation of Anticancer Activity of N,N-Dimethylaminomethylferrocene, Journal of Fundamental and Applied Sciences 11 (2019) 748–768. https://doi.org/10.4314/JFAS.V11I2.14.
L. Tabrizi, T.L.A. Nguyen, H.D.T. Tran, M.Q. Pham, D.Q. Dao, Antioxidant and Anticancer Properties of Functionalized Ferrocene with Hydroxycinnamate Derivatives—An Integrated Experimental and Theoretical Study, J Chem Inf Model 60 (2020) 6185–6203. https://doi.org/10.1021/acs.jcim.0c00730.
S. Bano, A. Khan, F. Asghar, M. Usman, A. Badshah, S. Ali, Computational and Pharmacological Evaluation of Ferrocene-Based Acyl Ureas and Homoleptic Cadmium Carboxylate Derivatives for Anti-diabetic Potential, Front Pharmacol 8 (2018) 1–16. https://doi.org/10.3389/fphar.2017.01001.
O. Domarle, G. Blampain, H. Agnaniet, T. Nzadiyabi, J. Lebibi, J. Brocard, L. Maciejewski, C. Biot, A.J. Georges, P. Millet, In Vitro Antimalarial Activity of a New Organometallic Analog, Ferrocene-Chloroquine, Antimicrob Agents Chemother 42 (1998) 540–544. https://doi.org/10.1128/AAC.42.3.540.
W.-Y. Guo, L.-Z. Chen, B.-N. Shen, X.-H. Liu, G.-P. Tai, Q.-S. Li, L. Gao, B.-F. Ruan, Synthesis and in vitro and in vivo anti-inflammatory activity of novel 4-ferrocenylchroman-2-one derivatives, J Enzyme Inhib Med Chem 34 (2019) 1678–1689. https://doi.org/10.1080/14756366.2019.1664499.
M. Milusheva, M. Todorova, V. Gledacheva, I. Stefanova, M. Feizi-Dehnayebi, M. Pencheva, P. Nedialkov, Y. Tumbarski, V. Yanakieva, S. Tsoneva, Novel anthranilic acid hybrids—An alternative weapon against inflammatory diseases, Pharmaceuticals 16 (2023) 1660.
A.S. Dorafshan Tabatabai, E. Dehghanian, H. Mansouri-Torshizi, M. Feizi-Dehnayebi, Computational and experimental examinations of new antitumor palladium (II) complex: CT-DNA-/BSA-binding, in-silico prediction, DFT perspective, docking, molecular dynamics simulation and ONIOM, J Biomol Struct Dyn 42 (2024) 5447–5469.
L. Messaadia, Y. Bekkar, M. Benamira, H. Lahmar, Predicting the antioxidant activity of some flavonoids of Arbutus plant: A theoretical approach, Chemical Physics Impact 1 (2020) 100007. https://doi.org/10.1016/j.chphi.2020.100007.
R. Bendaas, Y. Bekkar, L. Messaadia, L. Bourougaa, A. Messaoudi, S. Kiamouche, B. Messaoud, Computational-based investigation of antioxidative potential polyphenolic compounds of Salvia officinalis L.: combined DFT and molecular docking approaches, J Mol Model 30 (2024) 87. https://doi.org/10.1007/s00894-024-05866-8.
K. Karrouchi, S.A. Brandán, Y. Sert, H. El-Marzouqi, S. Radi, M. Ferbinteanu, M.E.A. Faouzi, Y. Garcia, Synthesis, X-ray structure, vibrational spectroscopy, DFT, biological evaluation and molecular docking studies of (E)-N’-(4-(dimethylamino) benzylidene)-5-methyl-1H-pyrazole-3-carbohydrazide, J Mol Struct 1219 (2020) 128541.
I.M. Khan, A. Khan, S. Shakya, M. Islam, Exploring the photocatalytic activity of synthesized hydrogen bonded charge transfer co-crystal of chloranilic acid with 2-ethylimidazole: DFT, molecular docking and spectrophotometric studies in different solvents, J Mol Struct 1277 (2023) 134862.
M. Alizadeh, Z. Mirjafary, H. Saeidian, Straightforward synthesis, spectroscopic characterizations and comprehensive DFT calculations of novel 1-ester 4-sulfonamide-1, 2, 3-triazole scaffolds, J Mol Struct 1203 (2020) 127405.
M.N. Tahir, K.S. Munawar, M. Feizi-Dehnayebi, M. Ashfaq, M.E. Muhammed, One-dimensional polymer of copper with salicylic acid and pyridine linkers: Synthesis, characterizations, solid state assembly investigation by hirshfeld surface analysis, and computational studies, J Mol Struct 1297 (2024) 136956.
F. Akman, A. Demirpolat, A.S. Kazachenko, A.S. Kazachenko, N. Issaoui, O. Al-Dossary, Molecular Structure, Electronic Properties, Reactivity (ELF, LOL, and Fukui), and NCI-RDG Studies of the Binary Mixture of Water and Essential Oil of Phlomis bruguieri, Molecules 28 (2023) 2684. https://doi.org/10.3390/molecules28062684.
S. Khan, H. Sajid, K. Ayub, T. Mahmood, Adsorption behaviour of chronic blistering agents on graphdiyne; excellent correlation among SAPT, reduced density gradient (RDG) and QTAIM analyses, J Mol Liq 316 (2020) 113860. https://doi.org/10.1016/j.molliq.2020.113860.
E.U.A.-R.-S.R.P. Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; Li, X.; Caricato, M.; Marenich, A.V.; Bloino, J., Janesko, B.G., Gomperts, R., Mennucci, B., Hratchian, H.P., Gaussian 16, Revision B.01, (2016) Gaussian 16, Revision A.03, Gaussian, Inc., Wallin.
A.D. Becke, Density‐functional thermochemistry. III. The role of exact exchange, J Chem Phys 98 (1993) 5648–5652. https://doi.org/10.1063/1.464913.
H.M. Berman, The Protein Data Bank, Nucleic Acids Res 28 (2000) 235–242. https://doi.org/10.1093/nar/28.1.235.
G.M. Morris, R. Huey, W. Lindstrom, M.F. Sanner, R.K. Belew, D.S. Goodsell, A.J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J Comput Chem 30 (2009) 2785–2791. https://doi.org/10.1002/jcc.21256.
S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B.A. Shoemaker, P.A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang, E.E. Bolton, PubChem 2023 update, Nucleic Acids Res 51 (2023) D1373–D1380. https://doi.org/10.1093/nar/gkac956.
D.S. BIOVIA, Discovery studio visualizer, 2020, Dassault Systèmes, San Diego (2020)., (n.d.).
R. Wang, Y. Lu, S. Wang, Comparative Evaluation of 11 Scoring Functions for Molecular Docking, J Med Chem 46 (2003) 2287–2303. https://doi.org/10.1021/jm0203783.
A. Sagaama, O. Noureddine, S.A. Brandán, A.J. Jędryka, H.T. Flakus, H. Ghalla, N. Issaoui, Molecular docking studies, structural and spectroscopic properties of monomeric and dimeric species of benzofuran-carboxylic acids derivatives: DFT calculations and biological activities, Comput Biol Chem 87 (2020) 107311. https://doi.org/10.1016/j.compbiolchem.2020.107311.
T. Ben Issa, A. Sagaama, N. Issaoui, Computational study of 3-thiophene acetic acid: Molecular docking, electronic and intermolecular interactions investigations, Comput Biol Chem 86 (2020) 107268. https://doi.org/10.1016/j.compbiolchem.2020.107268.
S. Ramalakshmi, K. Sonanki, R. Prakash, G. Usha, K. Karpagalakshmi, E.R. Nagarajan, N. Selvapalam, In-silico approach of effect of protein on complexation of cucurbit[7]uril with N- (ferrocenylmethyl) aniline, Mater Today Proc 51 (2021) 1733–1737. https://doi.org/10.1016/j.matpr.2020.11.720.
B. Kurt, H. Temel, M. Atlan, S. Kaya, Synthesis, characterization, DNA interaction and docking studies of novel Schiff base ligand derived from 2,6-diaminopyridine and its complexes, J Mol Struct 1209 (2020) 127928. https://doi.org/10.1016/j.molstruc.2020.127928.
A. Arunadevi, N. Raman, Indole-derived water-soluble N, O bi-dentate ligand-based mononuclear transition metal complexes: in silico and in vitro biological screening, molecular docking and macromolecule interaction studies, J Biomol Struct Dyn 38 (2020) 1499–1513. https://doi.org/10.1080/07391102.2019.1611475.
D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A.E. Mark, H.J.C. Berendsen, GROMACS: Fast, flexible, and free, J Comput Chem 26 (2005) 1701–1718. https://doi.org/10.1002/jcc.20291.
B. Lotfi, O. Mebarka, S.U. Khan, T.T. Htar, Pharmacophore-based virtual screening, molecular docking and molecular dynamics studies for the discovery of novel neuraminidase inhibitors, J Biomol Struct Dyn 42 (2024) 5308–5320. https://doi.org/10.1080/07391102.2023.2225007.
K. Anbarasu, S. Jayanthi, Identification of curcumin derivatives as human LMTK3 inhibitors for breast cancer: a docking, dynamics, and MM/PBSA approach, 3 Biotech 8 (2018) 228. https://doi.org/10.1007/s13205-018-1239-6.
Additional Files
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Journal of the Algerian Chemical Society

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

